Parametric simulation
(→Calorimeter) |
|||
| Line 1: | Line 1: | ||
| − | |||
==Talks== | ==Talks== | ||
Revision as of 16:10, 22 November 2021
Contents |
Talks
- Software meeting 2018-06-29, Friday, File:Sctau papas v2 20180629.pdf
- Workshop on future Super c-tau factories 2021-11-16 Parametric simulation of the SCT detector
SctParSim
A parametric simulation is a tool to receive a detector response without detailed description of interaction of particles with matter.
Implemented detector subsystems:
- drift chamber
- FARICH PID system
- calorimeter
- muon system
The parametric simulation yields the detector response in the SCT EDM format thus allowing to analyze its result in the same manner as the result of the full simulation.
How-to use the parametric simulation is demonstrated here
Detector configuration
The detector parameters can be changed in the run script (see Configuration section). The detector parameters and their default values can be viewed here.
SctParSimAlg
| Name to change parameter | Description | Default value |
|---|---|---|
| B | Detector magnetic field | 1.5 |
| mostProbMass | The mass of most probable particle | 0.13957 |
TrackSystemTool
| Name to change paramater | Description | Default value |
|---|---|---|
| trackRhoMin | Inner radius of barrel tracker, m | 0.1 |
| trackRhoMax | Outer radius of barrel tracker, m | 0.8 |
| trackZMin | Inner z coordinate of endcup tracker, m | 0 |
| trackZMax | Outer z coordinate of endcup tracker, m | 1 |
| trackMinPt | Minimum momentum, GeV | 0.05 |
| trackPtProb | Registration probabilities for different momentum, {GeV, prob} | {{0.1, 0.8}, {0.3, 0.9}, {1, 0.95), {10, 0.99}} |
| trackRadLen | Radiation length in the track system, m | 187 |
| trackResParPT | Parameterizaton parameters for xy projection | 0.00212 |
| trackResParPZ | Parameterization parameters for z projection | {0.001281, 0.00308} |
| trackLayerAx | The radius of layers anf the location radius of the anod layers, mm | {{6.306, 217.306}, {6.644, 227.1}, {7.165, 246.906}, {6.564, 341.938}, {6.794, 352.06}, {7.14, 371.992}, {7.388, 382.95}, {6.651, 467.57}, {6.823, 477.718}, {6.968, 488.097}, {7.12, 498.701}, {7.274, 509.535}, {6.768, 636.322}, {6.898, 646.501}, {7.007, 656.957}, {7.121, 667.581}, {6.791, 750.730}, {6.902, 761.027}, {6.995, 771.472}, {7.091, 782.061}} |
| trackLayerSt | The radius of layers and the location radius of the stereo layers, mm | {{6.473, 280.136}, {6.747, 290.136}, {7.182, 310.863}, {7.486, 321.938}, {6.603, 405.941}, {6.799, 416.04}, {7.104, 436.741}, {7.314, 447.606}, {6.741, 533.35}, {6.859, 543.615}, {7.026, 554.088, {7.161, 564.762}, {6.778, 584.801}, {6.919, 595.108}, {7.039, 605.606}, {7.163, 6169.289}, {6.746, 689.948}, {6.865, 700.185}, {7.041, 720.09}, {7.165, 730.775}} |
FARICHSystemTool
| Name to change paramater | Description | Default value |
|---|---|---|
| farichRhoMin | Inner radius of barrel FARICH system, m | 0.82 |
| farichRhoMax | Outer radius of barrel FARICH system, m | 0.9 |
| farichZMin | Inner z coordinate of endcup FARICH system, m | 1.02 |
| farichZMax | Outer z coordinate of encup FARICH system, m | 1.273 |
| farichHoleR | Hole radius of FARICH system | 0.3 |
| parSimFarichFileName | The path to the file with response histograms of FARICH | ./pi_ms_f1_mppc2_px3_d200_mla4_graph2d.root |
CaloSystemTool
| Name to change paramater | Description | Default value |
|---|---|---|
| caloRhoMin | Inner radius of barrel calorimeter, m | 1.09 |
| caloRhoMax | Outer radius of barrel calorimeter, m | 1.55 |
| caloZMin | Inner z coordinate of endcup calorimeter, m | 1.293 |
| caloZMax | Outer z coordinate of endcup calorimeter, m | 1.86 |
| caloCosthmax | Maximum cosine | 0.9 |
| caloClSize | Calorimeter cluster size, m | 0.045 |
| caloClSizeEGamma | Calorimeter cluster size for gamma, m | 0.15 |
| caloEMinBarrel | Minimal energy, GeV | 0.015 |
| caloEMinEndcup | Minimal energy, GeV | 0.015 |
| caloResPar | Parameterization parameters | {1.34e-2, 0.066e-2, 0.0, 0.82e-2} |
MuonSystemTool
| Name to change paramater | Description | Default value |
|---|---|---|
| muonRhoMin | Inner radius of barrel muon system, m | 1.87 |
| muonRhoMax | Outer radius of barrel muon system, m | 2.15 |
| muonZMin | Inner z coordinate of endcup muon system, m | 1.88 |
| muonZMax | Outer z coordinate of endcup muon system, m | 2.16 |
| parSimMuonFileNameMu | The path to the file with response histograms of muon system (muon) | ./g4beamline_mu_plus_100k_parse.root |
| parSimMuonFileNamePi | The path to the file with response histograms of muon system (pion) | ./g4beamline_pi_plus_100k_parse.root |
Parameterization
Drift chamber
The track resolution is described here.
The parametric simulation has two options in the tracker to the particle identification: an ionisation clusters counting (dNcl/dx) and dE/dx. The model of calculation the specific number of ionization clusters is taken from a TraPID option by F. Gracagnolo (The presentation on "Joint Workshop on Future tau-charm factory" in December 4--7, 2018). An energy losing is calculated using the resolution model from BaBar experiment. The The σ(dE/dx) is parameterized as
where α, β, γ is tuned on BaBar data.
FARICH PID system
The FARICH PID system works using the results of the full GEANT4 simulation. The system output is the particle speed and number of photons.
Calorimeter
The calorimeter resolution is sigma_E = e0 + e1 / E + e2 / sqrt(E) + e3 / E^0.25, the coefficients (e0, e1 etc.) are taken from the D. Epifanov presentation on Super C-Tau factory workshop in May 27, 2018.
There are two cluster sizes to gammas / electrons and other particles.
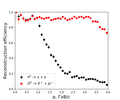
Reconstruction
The cross-linking data obtained by the track system and the calorimeter is implemented. This is implemented taking into account the geometric intersection of the calorimeter clusters.
The algorithm for the cross-linking data obtained by the track system and the calorimeter:
- union of geometrically intersecting calorimetric clusters
- finding a match between calorimetric clusters and tracks
- recalculating cluster characteristics (time, energy, cluster size, conversion point)
Muon system
The muon system works using the results of a reconducted stand-alone simulation on G4BeamLine. The system is a cylinder of eight absorber and sensitive polystyrene layers. The absorber is iron.
Output collection
The output ROOT-file contains:
- allGenParticles - MC particles
- pdgId
- charge
- vertex - x, y, and z coordinates
- p4 - momentum (px, py, pz) and mass
- Particles - the particle characteristics after the parametric simulation
- pdgId
- charge
- vertex - x, y, z coordinates
- p4 - measured momentum (px, py, pz) and the most probability particle mass
- dedx
- dedx_err
- dncldx - cluster counting
- dedxPid
- dncldxPid
- farichPid
- muPid
- TrackState - the helix characteristics (the track)
- phi
- theta
- qOverP
- d0
- z0
- referencePoint - x, y and z coordinates
- CaloClusters
- energy
- time
- position - x, y and z coordinates of the calorimeter entry point
- MuonHits
- layer - the last layer that registered a particle
- FARICHHits
- cellId
- energy
- time
- beta
- beta_err
- nphe0
- nphe
The more characteristics have intuitive names.
SctParSim (Python)
How to run
Login to stark or proxima machine.
ssh stark -X setupSCTAU; asetup SCTauSim,master,latest mkdir workarea cd workarea mkdir run cd run cp /home/razuvaev/public/misc/pi_ms_f1_mppc2_px3_d200_mla4_graph2d.root . cp /home/razuvaev/public/misc/gun1.cfg . cp /home/razuvaev/public/misc/g4beamline_pi_plus_100k_parse.root . cp /home/razuvaev/public/misc/g4beamline_mu_plus_100k_parse.root . cp /home/whitem/public/misc/pi_p.root . cp /home/whitem/public/misc/pi_m.root . cp /home/whitem/public/misc/mu_p.root . cp /home/whitem/public/misc/mu_m.root . runparsim.py
Options
| Option | Run example | Description |
|---|---|---|
-b, --batch
|
runparsim.py -b
|
Turn on the batch mode. Suppress EventDisplay execution. |
-c, --change
|
runparsim.py -c my_cfg_file.dat
|
Change some parameters of the detector. |
-n, --neve
|
runparsim.py -n 31415
|
The number of events to process. |
-g, --gun
|
runparsim.py -g
|
Turn on the particle gun mode. |
-ig, --input-gun
|
runparsim.py -g -ig my_gun.dat
|
Input particle gun configuration file. |
-i, --input
|
runparsim.py -i gen_dkpipi0.root
|
Specify the input file. |
-o, --output
|
runparsim.py -o parsim_dkpipi0.root
|
Specify the output file. |
--profile
|
runparsim.py --profile
|
Turn on the cProfile to analyse the performance. |
Event display
The event display has two projections: x-y and y-z. All detector subsystems are presented. Some of them overlay, especially PIDs, it is not important for parametric simulation, but let's study several options at once. All particles from AllGenParticles branch are presented at the plot by lines of different styles, colours and thicknesses. Somehow the line thickness corresponds to the particle mass, the wide line --- the more massive particle. Warm colours are devoted to positive charged particles and cold to negative ones. Also, particle lines are labeled.
The event display is switched on by default. To switch it off run simulation with -b option.
Detector configuration
The detector parameters can be changed via a configuration file my_cfg_file.dat placed in the main simulation folder. The file has a simple structure --- one parameter and its value(s) per line. A parameter's name and value should be separated by spaces. Arrays should be written in [] brackets. The values in arrays should be separated by commas. Empty lines and lines contained incorrectly parameter names are ignored.
In the example below the parameter at the first line and the second line is one number, while the parameter at the third line is an array.
trck.minPt 0.05 trck.corrMtx.pij -0.08 calo.resPar [0.167,0.0,0.011]
The parameters can be given in any order.
All detector parameters can be viewed in the configuration files:
| Subsystem | Congiguration file | Name to change parameters |
|---|---|---|
| ASHIPH | ashiphpars_std01.json
|
ashiph1030
|
| Calorimeter | calopars_std01.json
|
calo
|
| FARICH | farichpars_std01.json
|
farich
|
| FDIRC | fdircpars_std01.json
|
fdirc
|
| Muon system | muonpars_std01.json
|
muon
|
| ToF | tofpars_std01.json
|
tof
|
| ToP | toppars_std01.json
|
top
|
| Tracker | trkpars_std01.json
|
tracker
|
The detector subsystems sizes are stored in detlayout_std01.json. They can be changed in the same way.
Particle gun
Examples
Pictures
PAPAS (Old)
About papas, heppy et cetra
Particle propagation is done by geometry calculation. To valid the calculation several different cases were plotted.
Configure detector parameters
The detector parameters can be changed via a configuration file CTauPapas.cfg placed in the main papas simulation folder. The file has a simple structure --- one parameter and its value(s) per line. A parameter's name and value(s) should be separated by spaces. Empty lines and lines beginning with # are ignored.
In the example below the parameter at the first line is one number, while the parameter at the second line is an array.
ecal_emin_barrel 0.05
ecal_eres 1.34e-2 0.066e-2 0 0.82e-2
The parameters can be given in any order.
Configure detector parameters
The file ctau_input_sim.txt contains two lines. The first line is the path to a primary simulation file (see MC Data Sets page). The second line is an integer number of events to be processed.
How to run papas
Copy a directory with papas on stark the machine and go to this directory.
cd
cp -rf ~razuvaev/myheppy .
cd myheppy
There are a directory output for output files, detector configuration file CTauPapas.cfg, file ctau_input_sim.txt with a path to the file with primary generator events, and the folder heppy with heppy code itself. Let's go into it and tune environment.
cd heppy
source init.sh
Now it is time to run papas.
You may be asked a question because the output directory is not empty.
So just input y or clean the folder.
cd test
./heppy_loop.py ../../output/ ctau_cfg1.py
If it don't want to run try source ~razuvaev/.bashrc and source ../init.sh because it can be caused by the problem with environment variables.
When papas simulation has been done one need to present papas output to a suitable form and also add initial generator information.
cd ../../
./txt2tree.py
The output root tree is available in the file myheppy/output/txt2tree.root.
Output tree
The output tree contains branches which can be divided in several groups:
- reconstructed particle parameters;
- generated particle parameters;
- generated vertices;
- connection between reconstructed particles, generated particles and generated vertices.
The table below presents branches and description of their content.
| Name | Type | Length | Description |
|---|---|---|---|
| Reconstructed particles | |||
| n | int | 1 | The number of reconstructed particles. |
| px | float [] | n | The reconstructed particle momentum: x coordinate. |
| py | float [] | n | The reconstructed particle momentum: y coordinate. |
| pz | float [] | n | The reconstructed particle momentum: z coordinate. |
| Generated particles | |||
| n0 | int | 1 | The number of generated particles. |
| px0 | float [] | n0 | The generated particle momentum: x coordinate. |
| py0 | float [] | n0 | The generated particle momentum: y coordinate. |
| pz0 | float [] | n0 | The generated particle momentum: z coordinate. |
| Generated vertices | |||
| nv0 | int | 1 | The number of generated vertices. |
| vx0 | float [] | nv0 | The generated vertex: x coordinate. |
| vy0 | float [] | nv0 | The generated vertex: y coordinate. |
| vz0 | float [] | nv0 | The generated vertex: z coordinate. |
| Links | |||
| recgen | int [] | n | Transform a reconstructed particle index to the generated particle index. |
| genver | int [] | n0 | Transform a generated particle index to the generated vertex index. |
Analysis example
Here a short analysis example of
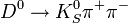 is presented.
The things are performed with PyROOT.
is presented.
The things are performed with PyROOT.
The data a taken from the available exclusive sample.
The code can be taken from github [1] or find at the stark cluster: /home/razuvaev/myheppy/search_dkspipi.py.
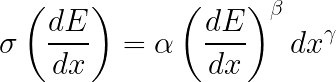




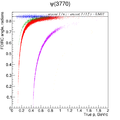


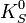 mass vs
mass vs 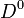 mass.
mass.
